Rous sarcoma virus
| Rous sarcoma virus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Ortervirales |
| Family: | Retroviridae |
| Genus: | Alpharetrovirus |
| Species: | Rous sarcoma virus
|
Rous sarcoma virus (RSV) (/raʊs/) is a retrovirus and is the first oncovirus to have been described. It causes sarcoma in chickens.
As with all retroviruses, it reverse transcribes its RNA genome into cDNA before integration into the host DNA.
History[edit]
RSV was discovered in 1911 by Peyton Rous, working at Rockefeller University in New York City, by injecting cell free extract of chicken tumour into healthy Plymouth Rock chickens. The extract was found to induce oncogenesis. The tumour was found to be composed of connective tissue (a sarcoma).[1][2] Thus, RSV became known as the first oncogenic retrovirus that could be used to study the development of cancer molecularly.[3]
In 1958, Harry Rubin and Howard Temin developed an assay where chicken embryo fibroblasts could be altered morphologically by RSV infection. Two years later Temin concluded that the transformed morphology of the cells was controlled by a genetic property of RSV. At that time it was unknown, but later the src gene was identified as responsible for morphological transformation in healthy cells. During the 1960s, two findings emerged: replication-competent isolated viruses were related to RSV, but were non-transforming, and an isolated replication-defective strain of RSV was transformation-competent. These two findings gave rise to the notion that viral replication and malignant transformation are separate processes in RSV.[4]
Rous was awarded the Nobel Prize in Physiology or Medicine for the significance of his discovery in 1966.[5] Subsequently, other oncogenic human viruses, such as Epstein–Barr virus, were discovered. Furthermore, oncogenes were found initially in retroviruses and then in cells.[3]
Structure and genome[edit]
| Retroviral 3'UTR stability element | |
|---|---|
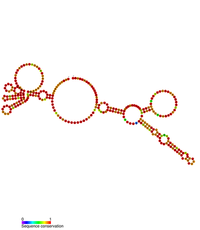 Predicted secondary structure of the Rous sarcoma virus retroviral 3'UTR stability element | |
| Identifiers | |
| Rfam | RF01417 |
| Other data | |
| RNA type | Cis-reg |
| PDB structures | PDBe |
RSV is a class VI enveloped virus with a positive sense RNA genome having a DNA intermediate.
Freely replicating RSV strains such as Prague-C have four genes:[6]
- gag – encodes capsid proteins
- pol – encodes reverse transcriptase
- env – encodes the envelope gene
- src – encodes a tyrosine kinase that attaches phosphate groups to the amino acid tyrosine in host cell proteins.
However, not all RSV strains have all four genes that allow it to both replicate on its own and cause transformation. It is not at all clear whether Rous's original strain was able to replicate on its own or if some descendants acquired this ability later.[7] Very few acutely oncogenic or transforming retroviruses are capable of replication without a helper virus, making non-defective strains of RSV quite unique.[8]
The RSV genome has terminal repeats enabling its integration into the host genome and also overexpression of RSV genes.
Src gene[edit]
The src gene is oncogenic as it triggers uncontrolled growth in abnormal host cells. It was the first retroviral oncogene to be discovered.[9] It is an acquired gene, found to be present throughout the animal kingdom with high levels of conservation between species.
The chicken src gene was taken up by RSV and incorporated into its genome.[10]
The v-Src gene is thought to confer the virus with the advantage of being able to stimulate uncontrolled mitosis of host cells, providing abundant cells for fresh infection.[11] The src gene is not essential for RSV reproduction but it greatly increases virulence when present.[9]
Src is a tyrosine kinase involved in regulation of cell growth and differentiation. It has an SH2 and SH3 domain, which are responsible for its activation and deactivation.[4]
RNA secondary structure[edit]
The RNA genome of RSV contains an extremely long 3' UTR that ranges between 5–7 kb in length which would usually direct it toward nonsense mediated decay (NMD) within the eukaryotic host cell. A conserved secondary structure element has been identified within the 3'UTR and is known as the Rous Sarcoma Virus Stability Element (RSE).[12] This element has been shown to prevent the degradation of the unspliced viral RNA.[12]
The RSE element was first identified in the genome of the Rous Sarcoma Virus but appears to be widely conserved across the avian retrovirus family. The RSE element is ~300 bp in length and located downstream of the gag natural translational termination codon. The secondary structure of the RSE element has been determined by RNAse digestion and SHAPE chemistry analysis.[13]
Other elements that have been identified in RSV include a primer binding site.[14]
Gag protein[edit]
Gag proteins are necessary for virion assembly and mature virus infection of the host cell. The gag protein (Pr76) for RSV contains 701 amino acids. It is cleaved by virus encoded protease, releasing products found in the infectious virion. These cleaved products include the matrix (MA), capsid (CA), and nucleocapsid (NC), which are able to enter other pathways to infect new cells.[15][16]
RSV envelope[edit]
RSV has an envelope which has one glycoprotein: env. Env is made up of gp85 and gp37, which are glycoproteins that assemble into oligomers. The function of env is to bind RSV to the host cell receptor and induce fusion with the target cell in a pH independent manner. The envelope is acquired during exocytosis. The virus buds or pushes on the plasma membrane, which allows it to leave the cell with a new outer membrane from the host cell.[15][17]
Replication cycle[edit]
Cell entry[edit]
There are two ways viruses can enter the host cell: cell receptor endocytosis or fusion. Fusion occurs when the virus fuses together with the target cell membrane and releases its genome into the cell. RSV enters the host cell through fusion of the host cell membrane.[18]
Transcription[edit]
In order for the RSV genome transcription to occur, a primer is required. 4S RNA is the primer for RSV and 70S RNA serves as the template for DNA synthesis. Reverse transcriptase, an RNA-dependent DNA polymerase, transcribes viral RNA into the full length DNA complement.[19]
References[edit]
- ^ Rous P (September 1910). "A Transmissible Avian Neoplasm (Sarcoma of the Common Fowl)". J. Exp. Med. 12 (5): 696–705. doi:10.1084/jem.12.5.696. PMC 2124810. PMID 19867354.
- ^ Rous P (April 1911). "A Sarcoma of the Fowl Transmissible by an Agent Separable from the Tumor Cells". J. Exp. Med. 13 (4): 397–411. doi:10.1084/jem.13.4.397. PMC 2124874. PMID 19867421.
- ^ Jump up to: a b Weiss RA, Vogt PK (November 2011). "100 years of Rous sarcoma virus". J. Exp. Med. 208 (12): 2351–5. doi:10.1084/jem.20112160. PMC 3256973. PMID 22110182.
- ^ Jump up to: a b Martin GS (June 2001). "The hunting of the Src". Nat. Rev. Mol. Cell Biol. 2 (6): 467–75. doi:10.1038/35073094. PMID 11389470. S2CID 205016442.
- ^ Nobelprize.org The Nobel Prize in Physiology or Medicine 1966: Peyton Rous, retrieved 1 Jul 2012
- ^ "Rous sarcoma virus - Prague C, complete genome". 26 July 2016.
- ^ Yatsula, B. A.; Geryk, J.; Briestanska, J.; Karakoz, I.; Svoboda, J.; Rynditch, A. V.; Calothy, G.; Dezelee, P. (1 October 1994). "Origin and evolution of the c-src-transducing avian sarcoma virus PR2257". Journal of General Virology. 75 (10): 2777–2781. doi:10.1099/0022-1317-75-10-2777. PMID 7931166.
The replication competent strains of Rous sarcoma virus (RSV) are exceptional in that they contain a complete set of replicative genes in addition to the transduced oncogene sequences. In this respect, they differ markedly from all other acutely transforming retroviruses, in which a large part of the replicative genes have been lost during transduction of cellular proto-oncogenes. However, since both the Bryan strain of RSV (Lerner & Hanafusa, 1984) and RSV29 (Dutta et al., 1985), the closest isolate to the original Rous tumour virus, lack the env gene, it is likely that the ancestral virus was also replication defective.
– See Vogt PK (2019) for discussion of whether PR2257 constitutes a de novo uptake. - ^ Vogt, PK (18 January 2019). "The Importance of Being Non-Defective: A Mini Review Dedicated to the Memory of Jan Svoboda". Viruses. 11 (1): 80. doi:10.3390/v11010080. PMC 6360021. PMID 30669277.
- ^ Jump up to: a b Vogt PK (September 2012). "Retroviral oncogenes: a historical primer". Nat. Rev. Cancer. 12 (9): 639–48. doi:10.1038/nrc3320. PMC 3428493. PMID 22898541.
- ^ Swanstrom, R; Parker, R C; Varmus, H E; Bishop, J M (May 1983). "Transduction of a cellular oncogene: the genesis of Rous sarcoma virus". Proceedings of the National Academy of Sciences. 80 (9): 2519–2523. Bibcode:1983PNAS...80.2519S. doi:10.1073/pnas.80.9.2519. PMC 393857. PMID 6302692.
- ^ Wyke, AW; Frame, MC; Gillespie, DA; Chudleigh, A; Wyke, JA (October 1995). "Mitogenesis by v-Src: fluctuations throughout G1 of classical immediate early AP-1 and mitogen-activated protein kinase responses that parallel the need for the oncoprotein". Cell Growth & Differentiation. 6 (10): 1225–34. PMID 8845299.
- ^ Jump up to: a b Weil JE, Beemon KL (January 2006). "A 3' UTR sequence stabilizes termination codons in the unspliced RNA of Rous sarcoma virus". RNA. 12 (1): 102–10. doi:10.1261/rna.2129806. PMC 1370890. PMID 16301601.
- ^ Weil JE, Hadjithomas M, Beemon KL (March 2009). "Structural characterization of the Rous sarcoma virus RNA stability element". J. Virol. 83 (5): 2119–29. doi:10.1128/JVI.02113-08. PMC 2643715. PMID 19091866.
- ^ Johnson M, Morris S, Chen A, Stavnezer E, Leis J (2004). "Selection of functional mutations in the U5-IR stem and loop regions of the Rous sarcoma virus genome". BMC Biol. 2: 8. doi:10.1186/1741-7007-2-8. PMC 428589. PMID 15153244.
- ^ Jump up to: a b Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J (October 1994). "An assembly domain of the Rous sarcoma virus Gag protein required late in budding". J. Virol. 68 (10): 6605–18. doi:10.1128/JVI.68.10.6605-6618.1994. PMC 237081. PMID 8083996.
- ^ Nadaraia-Hoke S, Bann DV, Lochmann TL, Gudleski-O'Regan N, Parent LJ (March 2013). "Alterations in the MA and NC domains modulate phosphoinositide-dependent plasma membrane localization of the Rous sarcoma virus Gag protein". J. Virol. 87 (6): 3609–15. doi:10.1128/JVI.03059-12. PMC 3592118. PMID 23325682.
- ^ Einfeld D, Hunter E (November 1988). "Oligomeric structure of a prototype retrovirus glycoprotein". Proc. Natl. Acad. Sci. U.S.A. 85 (22): 8688–92. Bibcode:1988PNAS...85.8688E. doi:10.1073/pnas.85.22.8688. PMC 282525. PMID 2847170.
- ^ Gilbert JM, Mason D, White JM (October 1990). "Fusion of Rous sarcoma virus with host cells does not require exposure to low pH". J. Virol. 64 (10): 5106–13. doi:10.1128/JVI.64.10.5106-5113.1990. PMC 248002. PMID 2168989.
- ^ Dahlberg JE, Sawyer RC, Taylor JM, Faras AJ, Levinson WE, Goodman HM, Bishop JM (May 1974). "Transcription of DNA from the 70S RNA of Rous sarcoma virus. I. Identification of a specific 4S RNA which serves as primer". J. Virol. 13 (5): 1126–33. doi:10.1128/JVI.13.5.1126-1133.1974. PMC 355423. PMID 4132919.
External links[edit]
- the Rous sarcoma virus (RSV) at rcn.com Archived 2006-04-29 at the Wayback Machine
- molecular expressions at magnet.fsu.edu Archived 2006-02-13 at the Wayback Machine
- Rous+sarcoma+virus at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Rfam entry for retroviral 3'UTR stability element
