Parvoviridae
| Parvoviridae | |
|---|---|
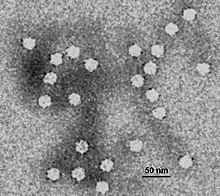
| |
| Electron micrograph of canine parvovirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Quintoviricetes |
| Order: | Piccovirales |
| Family: | Parvoviridae |
| Genera | |
Parvoviruses are a family of animal viruses that constitute the family Parvoviridae. They have linear, single-stranded DNA (ssDNA) genomes that typically contain two genes encoding for a replication initiator protein, called NS1, and the protein the viral capsid is made of. The coding portion of the genome is flanked by telomeres at each end that form into hairpin loops that are important during replication. Parvovirus virions are small compared to most viruses, at 23–28 nanometers in diameter, and contain the genome enclosed in an icosahedral capsid that has a rugged surface.
Parvoviruses enter a host cell by endocytosis, travelling to the nucleus where they wait until the cell enters its replication stage. At that point, the genome is uncoated and the coding portion is replicated. Viral messenger RNA (mRNA) is then transcribed and translated, resulting in NS1 initiating replication. During replication, the hairpins repeatedly unfold, are replicated, and refold to change the direction of replication to progress back and forth along the genome in a process called rolling hairpin replication that produces a molecule containing numerous copies of the genome. Progeny ssDNA genomes are excised from this concatemer and packaged into capsids. Mature virions leave the cell by exocytosis or lysis.
Parvoviruses are believed to be descended from ssDNA viruses that have circular genomes that form a loop because these viruses encode a replication initiator protein that is related to NS1 and have a similar replication mechanism. Another group of viruses called bidnaviruses appear to be descended from parvoviruses. Within the family, three subfamilies, 26 genera, and 126 species are recognized. Parvoviridae is the sole family in the order Piccovirales, which is the sole order in the class Quintoviricetes. This class is assigned to the phylum Cossaviricota, which also includes papillomaviruses, polyomaviruses, and bidnaviruses.
A variety of diseases in animals are caused by parvoviruses. Notably, the canine parvovirus and feline parvovirus cause severe disease in dogs and cats, respectively. In pigs, the porcine parvovirus is a major cause of infertility. Human parvoviruses are less severe, the two most notable being parvovirus B19, which causes a variety of illnesses including fifth disease in children, and human bocavirus 1, which is a common cause of acute respiratory tract illness, especially in young children. In medicine, recombinant adeno-associated viruses (AAV) have become an important vector for delivering genes to the cell nucleus during gene therapy.
Animal parvoviruses were first discovered in the 1960s, including minute virus of mice, which is frequently used to study parvovirus replication. Many AAVs were also discovered during this time period and research on them over time has revealed their benefit as a form of medicine. The first pathogenic human parvovirus to be discovered was parvovirus B19 in 1974, which became associated with various diseases throughout the 1980s. Parvoviruses were first classified as the genus Parvovirus in 1971 but were elevated to family status in 1975. They take their name from the Latin word parvum, meaning 'small' or 'tiny', referring to the small size of the virus's virions.
Genome
[edit]Parvoviruses have linear, single-stranded DNA (ssDNA) genomes that are about 4–6 kilobases (kb) in length. The parvovirus genome typically contains two genes, termed the NS/rep gene and the VP/cap gene.[1] The NS gene encodes the non-structural (NS) protein NS1, which is the replication initiator protein, and the VP gene encodes the viral protein (VP) that the viral capsid is made of. NS1 contains an HUH superfamily endonuclease domain near its N-terminus, containing both site-specific binding activity and site-specific nicking activity, and a superfamily 3 (SF3) helicase domain toward the C-terminus. Most parvoviruses contain a transcriptional activation domain near the C-terminus that upregulates transcription from viral promoters as well as alternate or overlapping open reading frames that encode a small number of supporting proteins involved in different aspects of the viral life cycle.[2]
The coding portion of the genome is flanked at each end by terminal sequences about 116–550 nucleotides (nt) in length that consist of imperfect palindromes folded into hairpin loop structures. These hairpin loops contain most of the cis-acting information required for DNA replication and packaging and act as hinges during replication to change the direction of replication. When the genome is converted to double-stranded forms, replication origin sites are created involving sequences in and adjacent to the hairpins.[2][3]
Genomic DNA strands in mature virions may be positive-sense or negative-sense. This varies from species to species as some have a preference for packaging strands of one polarity, others package varying proportions, and others package both sense strands at equal proportions. These preferences reflect the efficiency with which progeny strands are synthesized, which in turn reflects the efficiency of specific replication origin sites.[2] The 3′-end (usually pronounced "three prime end") of a negative sense strand, and the 5′-end (usually pronounced "five prime end") of a positive sense strand, is called the left end, and the 5′-end of the negative sense strand, and the 3′-end of a positive sense strand, is called the right end.[2][4][5]
Structure
[edit]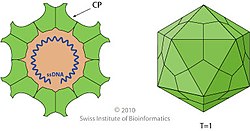

Parvovirus virions are 23–28 nanometers (nm) in diameter and consist of the genome enclosed inside a capsid that is icosahedral in shape with a rugged surface. The capsid is composed of 60 structurally equivalent polypeptide chains derived from the C-terminal end of a VP protein's sequence, interlocking extensively to form an icosahedron with 60 asymmetric, superficial triangular units. These units have 3-fold radial symmetry at two vertices and 5-fold radial symmetry at one, with 2-fold radial symmetry at the line opposite of the 5-fold vertex and a 2/5 circular fold wall surrounding the point of the 5-fold vertex. Twenty 3-fold vertices, thirty 2-fold lines, and twelve 5-fold vertices exist per capsid, the latter corresponding to the 12 vertices of the icosahedron.[2]
Typical features of the capsid surface include depressions at each 2-fold axis, elevated protrusions surrounding the 3-fold axes, and raised cylindrical projections made of five beta-barrels[6] surrounded by canyon-like depressions at the 5-fold axes. Each of these cylinders potentially contains an opening to connect the exterior of the capsid to the interior, which mediates entry and exit of the genome. About 20 nucleotides from the 5′-end of the genome may remain exposed outside of the capsid carrying a copy of NS1 bound to the 5′-end, which is a result of how the genome is synthesized and packaged.[2]
Varying sizes of the VP protein are expressed for different parvoviruses, the smaller ones, VP2–5, being expressed at a higher frequency than the large size, VP1. The smaller VPs share a common C-terminus with different N-terminus lengths due to truncation. For VP1, the N-terminus is extended to contain regions important in the replication cycle, and it is incorporated into the capsid, typically 5–10 per capsid, with the common C-terminus responsible for assembling capsids.[1][2]
Each VP monomer contains a core beta-barrel structure called the jelly roll motif of eight strands arranged in two adjacent antiparallel beta sheets, labeled CHEF and BIDG after the individual strands, the latter forming the interior surface of the capsid. Individual beta strands are connected by loops that have varying length, sequence, and conformation, and most of these loops extend toward the exterior surface, giving parvoviruses their unique, rough surface. Related parvoviruses share their surface topologies and VP protein folds to a greater degree than their sequence identities, so the structure of the capsid and capsid protein are useful indicators of phylogeny.[1][2]
Life cycle
[edit]Parvoviruses enter cells by endocytosis, using a variety of cellular receptors to bind to the host cell. In endosomes, many parvoviruses undergo a change in conformation so that the phospholipase A2 (PLA2) domain on the VP1 N-termini are exposed so the virion can penetrate lipid bilayer membranes. Intracellular trafficking of virions varies, but virions ultimately arrive to the nucleus, inside of which the genome is uncoated from the capsid. Based on studies of minute virus of mice (MVM), the genome is ejected from the capsid in a 3′-to-5′ direction from one of the openings in the capsid, leaving the 5′-end of the DNA attached to the capsid.[2]
Parvoviruses lack the ability to induce cells into their DNA replication stage, called S-phase, so they must wait in the nucleus until the host cell enters S-phase on its own. This makes cell populations that divide rapidly, such as fetal cells, an excellent environment for parvoviruses. Adeno-associated viruses (AAV) are dependent on helper viruses, which may be an adenovirus or a herpesvirus, since coinfection alters the cellular environment to allow for replication.[2] In the absence of coinfection, AAV's genome is integrated into the host cell's genome until coinfection occurs.[7] Infected cells that enter S-phase are forced to synthesize viral DNA and cannot leave S-phase. Parvoviruses establish replication foci in the nucleus that grow progressively larger as infection progresses.[8]
Once a cell enters S-phase and the genome is uncoated, a host DNA polymerase uses the 3′-end of the 3′ hairpin as a primer to synthesize a complementary DNA strand for the coding portion of the genome, which is connected to the 5′-end of the 5′ hairpin.[3][7][9] Messenger RNA (mRNA) that encodes NS1 is then transcribed from the genome by the DNA polymerase, capped and polyadenylated, and translated by host ribosomes to synthesize NS1.[2][5][10] If proteins are encoded in multiple co-linear frames, then alternative splicing, suboptimal translation initiation, or leaky scanning may be used to translate different gene products.[2]
Parvoviruses replicate their genome via rolling hairpin replication, a unidirectional, strand displacement form of DNA replication that is initiated by NS1. Replication begins once NS1 binds to and makes a nick in a replication origin site in the duplex DNA molecule at the end of one hairpin. Nicking releases the 3′-end of the nicked strand as a free hydroxyl (-OH) to prime DNA synthesis[2] with NS1 remaining attached to the 5′-end.[7] The nick causes the adjacent hairpin to unfold into a linear, extended form. At the 3′-OH, a replication fork is established using NS1's helicase activity, and the extended telomere is replicated by the DNA polymerase.[10][11] The two telomere strands then refold back in on themselves to their original configurations, which repositions the replication fork to switch templates to the other strand and move in the opposite direction toward the other end of the genome.[12][13]
Parvoviruses vary in whether the termini are similar or the same, called homotelomeric parvoviruses, or different, called heterotelomeric parvoviruses. In general, homotelomeric parvoviruses, such as AAV and B19, replicate both ends of their genome through the aforementioned process, called terminal resolution, and their hairpin sequences are contained within larger (inverted) terminal repeats. Heterotelomeric viruses, such as minute virus of mice (MVM), replicate one end by terminal resolution and the other end via an asymmetric process called junction resolution[2][14] so that the correct orientation of the telomere can be copied.[15]
During asymmetric junction resolution, the duplex extended-form telomeres refold in on themselves into a cruciform shape. A replication origin site on the lower strand of the right arm of the cruciform is nicked by NS1, leading to the lower arm of the cruciform unfolding into its linear extended form. A replication fork established at the nick site moves down the extended lower arm to copy the lower arm's sequence. The two strands of the lower arm then refold to reposition the replication fork to go back toward the other end, displacing the upper strand in the process.[16]
The back and forth, end-to-end pattern of rolling hairpin replication produces a concatemer containing multiple copies of the genome.[2][3] NS1 periodically makes nicks in this molecule and, through a combination of terminal resolution and junction resolution, individual strands of the genome are excised from the concatemer.[9][13] Excised genomes may either be recycled for further rounds of replication or packaged into progeny capsids.[7] Translation of mRNA containing VP proteins leads to the accumulation of capsid proteins in the nucleus that assemble into these empty capsids.[8]
Genomes are encapsidated at one of the capsid's vertices through a portal,[2] potentially the one opposite the portal used to expel the genome.[5] Once complete virions have been constructed, they may be exported from the nucleus to the exterior of the cell before disintegration of the nucleus. Disruption of the host cell environment may also occur later on in the infection. This results in cell lysis via necrosis or apoptosis, which releases virions to the outside of the cell.[2][8]
Evolution
[edit]Parvoviruses are believed to be descended from ssDNA viruses that have a circular genome that forms a loop and which replicate via rolling circle replication, which is similar to rolling hairpin replication. These circular ssDNA viruses encode a replication initiator protein that is related to and possesses many of the same characteristics as the replication initiator protein of parvoviruses, such as the HUH endonuclease domain and the SF3 helicase domain.[17] In contrast to these other replication initiator proteins, NS1 shows only vestigial traces of being able to perform ligation, which is a key part of rolling circle replication.[8] The Bidnaviridae family, which are also linear ssDNA viruses, appear to be descended from a parvovirus that had its genome integrated into the genome of a polinton, a type of DNA transposon related to viruses in the realm Varidnaviria.[17]
Based on phylogenetic analysis of the SF3 helicase, parvoviruses split into two branches early in their evolutionary history, one of which contains viruses assigned to the subfamily Hamaparvovirinae. The other branch split into two sublineages that constitute the other two subfamilies, Densovirinae and Parvovirinae.[18] Parvoviruses in the Hamaparvovirinae lineage are likely all heterotelomeric, Densovirinae are exclusively homotelomeric, and Parvovirinae varies.[2] Telomere sequences have significant complexity and diversity, suggesting that many species have co-opted them to perform additional functions.[7][10] Parvoviruses are also considered to have high rates of genetic mutations and recombinations.[2][9]
Classification
[edit]Parvoviruses constitute the family Parvoviridae. The family is the sole family in the order Piccovirales, which is the sole order in the class Quintoviricetes. The class Quintoviricetes belongs to the phylum Cossaviricota, which also includes papillomaviruses, polyomaviruses, and bidnaviruses. Cossaviricota is included in the kingdom Shotokuvirae, which is assigned to the realm Monodnaviria. Parvoviridae belongs to Group II: ssDNA viruses in the Baltimore classification system, which groups viruses together based on their manner of mRNA synthesis. Within Parvoviridae, three subfamilies, 26 genera, and 126 species are recognized as of 2020 (-virinae denotes subfamily and -virus denotes genus):[18][19]
- Densovirinae (11 genera, 21 species)
- Aquambidensovirus (3 species)
- Blattambidensovirus (1 species)
- Diciambidensovirus (1 species)
- Hemiambidensovirus (2 species)
- Iteradensovirus (5 species)
- Miniambidensovirus (1 species)
- Muscodensovirus (1 species)
- Pefuambidensovirus (1 species)
- Protoambidensovirus (2 species)
- Scindoambidensovirus (3 species)
- Tetuambidensovirus (1 species)
- Hamaparvovirinae (5 genera, 21 species)
- Brevihamaparvovirus (2 species)
- Chaphamaparvovirus (16 species)
- Hepanhamaparvovirus (1 species)
- Ichthamaparvovirus (1 species)
- Penstylhamaparvovirus (1 species)
- Parvovirinae (10 genera, 84 species)
- Amdoparvovirus (5 species)
- Artiparvovirus (1 species)
- Aveparvovirus (3 species)
- Bocaparvovirus (28 species)
- Copiparvovirus (7 species)
- Dependoparvovirus (11 species)
- Erythroparvovirus (7 species)
- Loriparvovirus (1 species)
- Protoparvovirus (15 species)
- Tetraparvovirus (6 species)
Parvoviruses are assigned to the same species if they share at least 85% of their protein sequence identities. Species are grouped together in a genus based on phylogeny of the NS1 and SF3 helicase domains, as well as similarity of NS1 sequence identity and coverage. If these criteria aren't satisfied, then genera can still be established provided that common ancestry is supported. The three subfamilies are distinguished based on phylogeny of the SF3 helicase domain, which corresponds to host range: viruses in Densovirinae infect invertebrates, viruses in Hamaparvovirinae infect invertebrates and vertebrates, and viruses in Parvovirinae infect vertebrates.[18]
Disease
[edit]
In humans, the most prominent parvoviruses that cause disease are parvovirus B19 and human bocavirus 1. B19 infection is often asymptomatic but can manifest in a variety of ways, including Fifth disease with its characteristic rash in children, persistent anemia in immunocompromised persons and in people who have underlying hemoglobinopathies,[20] transient aplastic crises, hydrops fetalis in pregnant women, and arthropathy. Human bocavirus 1 is a common cause of acute respiratory tract infection, especially in young children, wheezing being a common symptom. Other parvoviruses associated with different diseases in humans include human parvovirus 4 and human bufavirus, though the manner by which these viruses cause disease is unclear.[6]
Carnivore-infecting viruses in the genus Protoparvovirus, in contrast to human parvoviruses, are more life-threatening.[2] Canine parvovirus causes severe illness in dogs, the most common symptom being hemorrhagic enteritis, with up to a 70% mortality rate in pups but usually less than 1% in adults.[21] Feline parvovirus, a closely related virus,[22] likewise causes severe illness in cats along with panleukopenia.[23][24] In pigs, porcine parvovirus is a major cause of infertility as infection frequently leads to death of the fetus.[25]
Use in medicine
[edit]Adeno-associated viruses have become an important vector for gene therapy aimed at treating genetic diseases, such as those caused by a single mutation. The recombinant AAV (rAAV) contains a viral capsid but lacks a complete viral genome. Instead, the typical nucleic acid packaged into the capsid contains a promoter region, the gene of interest, and a terminator region, all contained within two inverted terminal repeats derived from the viral genome. rAAV essentially acts as a container that can traverse the cell membrane and deliver its nucleic acid cargo to the nucleus.[26][27]
History
[edit]Parvoviruses were discovered relatively late in comparison to other prominent virus families, potentially due to their small size. In the late 1950s[28] and 1960s,[29] a variety of animal parvoviruses were discovered, including minute virus of mice,[30] which has since been used extensively to study rolling hairpin replication.[31] Many AAVs were also discovered during this time period[32] and research on them led to their first usage in gene therapy in the 1980s. Over time, improvements in aspects such as vector design led to certain AAV gene therapy products reaching clinical efficacy in 2008 and being approved in the following years.[27]
In 1974, the first pathogenic human parvovirus was discovered by Yvonne Cossart, et al. When testing for the hepatitis B virus's surface antigen, one serum sample gave anomalous results and with electron microscopy was shown to contain a virus resembling animal parvoviruses. This virus was named B19 after the coding of the serum sample, number 19 in panel B.[20][33] B19 was later recognized as a species by the International Committee on Taxonomy of Viruses (ICTV) in 1985, and throughout the 1980s it increasingly became associated with various diseases.[33]
In the ICTV's first report in 1971, parvoviruses were grouped together in the genus Parvovirus.[30][32] They were elevated to the rank of family in 1975 and remained unassigned to higher taxa until 2019, when they were assigned to higher taxa up to the highest rank, realm.[34] The family was reorganized in 2019, departing from the "traditional" invertebrate-vertebrate distinction between Densovirinae and Parvovirinae and instead distinguishing the subfamilies based on helicase phylogeny, leading to the establishment of a new subfamily, Hamaparvovirinae.[18]
Etymology
[edit]Parvoviruses take their name from Latin parvus or parvum, meaning small or tiny, referring to the small size of parvovirus virions compared to most other viruses.[2][20] In the family name Parvoviridae, -viridae is the suffix used for virus families.[35] The order Piccovirales takes the first part of its name from the Italian word piccolo, meaning small, and the second part is the suffix used for virus orders. The class Quintoviricetes takes the first part of its name from the Galician word quinto, meaning fifth, referring to fifth disease (erythema infectiosum) caused by parvovirus B19, and viricetes, the suffix used for virus classes.[17]
See also
[edit]Citations
[edit]- ^ a b c Mietzsch M, Pénzes JJ, Agbandje-McKenna M (20 April 2019). "Twenty-Five Years of Structural Parvovirology". Viruses. 11 (4): 362. doi:10.3390/v11040362. PMC 6521121. PMID 31010002.
- ^ a b c d e f g h i j k l m n o p q r s t u Cotmore SF, Agbandje-McKenna M, Canuti M, Chiorini JA, Eis-Hubinger AM, Hughes J, Mietzsch M, Modha S, Ogliastro M, Pénzes JJ, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P (March 2019). "ICTV Virus Taxonomy Profile: Parvoviridae". J Gen Virol. 100 (3): 367–368. doi:10.1099/jgv.0.001212. PMC 6537627. PMID 30672729. Retrieved 24 January 2021.
- ^ a b c Kerr, Cotmore & Bloom 2005, p. 177.
- ^ Kerr, Cotmore & Bloom 2005, p. 172.
- ^ a b c Cotmore SF, Tattersall P (1 February 2013). "Parvovirus diversity and DNA damage responses". Cold Spring Harb Perspect Biol. 5 (2): a012989. doi:10.1101/cshperspect.a012989. PMC 3552509. PMID 23293137.
- ^ a b Qiu J, Söderlund-Venermo M, Young NS (January 2017). "Human Parvoviruses". Clin Microbiol Rev. 30 (1): 43–113. doi:10.1128/CMR.00040-16. PMC 5217800. PMID 27806994.
- ^ a b c d e Cotmore SF, Tattersall P (1996). "Parvovirus DNA replication" (PDF). Cold Spring Harbor Monograph Archive. 31: 799–813. doi:10.1101/0.799-813 (inactive 31 January 2024). Retrieved 24 January 2021.
{{cite journal}}: CS1 maint: DOI inactive as of January 2024 (link) - ^ a b c d Kerr, Cotmore & Bloom 2005, p. 175.
- ^ a b c Martin DP, Biagini P, Lefeuvre P, Golden M, Roumagnec P, Varsani A (September 2011). "Recombination in eukaryotic single stranded DNA viruses". Viruses. 3 (9): 1699–1738. doi:10.3390/v3091699. PMC 3187698. PMID 21994803.
- ^ a b c Kerr, Cotmore & Bloom 2005, p. 173.
- ^ Kerr, Cotmore & Bloom 2005, p. 180.
- ^ Kerr, Cotmore & Bloom 2005, p. 179.
- ^ a b Kerr, Cotmore & Bloom 2005, p. 181.
- ^ Kerr, Cotmore & Bloom 2005, p. 171–172, 177, 179.
- ^ Kerr, Cotmore & Bloom 2005, p. 182.
- ^ Kerr, Cotmore & Bloom 2005, p. 182–184.
- ^ a b c Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH (18 October 2019). "Create a megataxonomic framework, filling all principal taxonomic ranks, for ssDNA viruses" (docx). ICTV. Retrieved 24 January 2021.
- ^ a b c d Penzes JJ, Soderlund-Venermo M, Canuti M, Eis-Huebinger AM, Hughes J, Cotmore SF. "Re-organize the family Parvoviridae" (docx). ICTV. Retrieved 24 January 2021.
- ^ "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 10 May 2021.
- ^ a b c Fonseca EK (February 2018). "Etymologia: Parvovirus". Emerg Infect Dis. 24 (2): 293. doi:10.3201/eid2402.ET2402. PMC 5782889.
- ^ Decaro N, Buonavoglia C (24 February 2012). "Canine parvovirus--a review of epidemiological and diagnostic aspects, with emphasis on type 2c". Vet Microbiol. 155 (1): 1–12. doi:10.1016/j.vetmic.2011.09.007. PMC 7173204. PMID 21962408.
- ^ Cotmore SF, McKenna MA, Chiorini JA, Gatherer D, Mukha DV, Pintel DJ, Qiu J, Soderland-Venermo M, Tattersall P, Tijssen P. "Rationalization and extension of the taxonomy of the family Parvoviridae" (PDF). ICTV. Retrieved 24 January 2021.
- ^ Parrish CR (March 1995). "Pathogenesis of feline panleukopenia virus and canine parvovirus". Baillière's Clin Haematol. 8 (1): 57–71. doi:10.1016/s0950-3536(05)80232-x. PMC 7134857. PMID 7663051.
- ^ "Feline panleukopenia". American Veterinary Medical Association. Retrieved 24 January 2021.
- ^ Mészáros I, Olasz F, Cságola A, Tijssen P, Zádori Z (20 December 2017). "Biology of Porcine Parvovirus (Ungulate parvovirus 1)". Viruses. 9 (12): 393. doi:10.3390/v9120393. PMC 5744167. PMID 29261104.
- ^ Naso MF, Tomkowicz B, Perry WL, Strohl WR (August 2017). "Adeno-Associated Virus (AAV) as a Vector for Gene Therapy". BioDrugs. 31 (4): 317–334. doi:10.1007/s40259-017-0234-5. PMC 5548848. PMID 28669112.
- ^ a b Wang D, Tai PW, Gao G (May 2019). "Adeno-associated virus vector as a platform for gene therapy delivery". Nat Rev Drug Discov. 18 (5): 358–378. doi:10.1038/s41573-019-0012-9. PMC 6927556. PMID 30710128.
- ^ Kilham L, Olivier LJ (April 1959). "A latent virus of rats isolated in tissue culture". Virology. 7 (4): 428–437. doi:10.1016/0042-6822(59)90071-6. PMID 13669314.
- ^ "Parvovirus". Stanford University. Retrieved 24 January 2021.
- ^ a b "ICTV Taxonomy history: Rodent protoparvovirus 1". ICTV. Retrieved 24 January 2021.
- ^ Kerr, Cotmore & Bloom 2005, p. 171–185.
- ^ a b "ICTV Taxonomy history: Adeno-associated dependoparvovirus A". ICTV. Retrieved 24 January 2021.
- ^ a b Heegaard ED, Brown KE (July 2002). "Human parvovirus B19". Clin Microbiol Rev. 15 (3): 485–505. doi:10.1128/cmr.15.3.485-505.2002. PMC 118081. PMID 12097253.
- ^ "ICTV Taxonomy history: Parvoviridae". ICTV. Retrieved 24 January 2021.
- ^ "ICTV Code". ICTV. Retrieved 24 January 2021.
General and cited references
[edit]- Kerr J, Cotmore S, Bloom ME (25 November 2005). Parvoviruses. CRC Press. pp. 171–185. ISBN 9781444114782.
External links
[edit] Media related to Parvoviridae at Wikimedia Commons
Media related to Parvoviridae at Wikimedia Commons Data related to Parvoviridae at Wikispecies
Data related to Parvoviridae at Wikispecies

