Apomorphy and synapomorphy
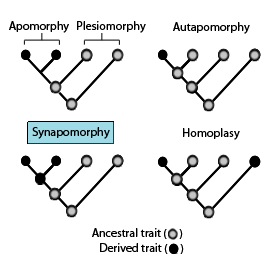
In phylogenetics, an apomorphy (or derived trait) is a novel character or character state that has evolved from its ancestral form (or plesiomorphy).[2][3][4] A synapomorphy is an apomorphy shared by two or more taxa and is therefore hypothesized to have evolved in their most recent common ancestor.[1][5][3][6][7][8][9] In cladistics, synapomorphy implies homology.[5]
Examples of apomorphy are the presence of erect gait, fur, the evolution of three middle ear bones, and mammary glands in mammals but not in other vertebrate animals such as amphibians or reptiles, which have retained their ancestral traits of a sprawling gait and lack of fur.[10] Thus, these derived traits are also synapomorphies of mammals in general as they are not shared by other vertebrate animals.[10]
Etymology
[edit]The word synapomorphy—coined by German entomologist Willi Hennig—is derived from the Ancient Greek words σύν (sún), meaning "with, together"; ἀπό (apó), meaning "away from"; and μορφή (morphḗ), meaning "shape, form".
Examples
[edit]Lampreys and sharks share features like gills and cartilaginous skeletons, but these traits are also present in many invertebrates. In contrast, the presence of jaws and paired appendages[11] in both sharks and cows, but not in lampreys or close invertebrate relatives, identifies these traits as synapomorphies. This supports the hypothesis that cows and sharks are more closely related to each other than to lampreys.
Clade analysis
[edit]The concept of synapomorphy depends on a given clade in the tree of life. Cladograms are diagrams that depict evolutionary relationships within groups of taxa. These illustrations are accurate predictive device in modern genetics. They are usually depicted in either tree or ladder form. Synapomorphies then create evidence for historical relationships and their associated hierarchical structure. Evolutionarily, a synapomorphy is the marker for the most recent common ancestor of the monophyletic group consisting of a set of taxa in a cladogram.[12] What counts as a synapomorphy for one clade may well be a primitive character or plesiomorphy at a less inclusive or nested clade. For example, the presence of mammary glands is a synapomorphy for mammals in relation to tetrapods but is a symplesiomorphy for mammals in relation to one another—rodents and primates, for example. So the concept can be understood as well in terms of "a character newer than" (autapomorphy) and "a character older than" (plesiomorphy) the apomorphy: mammary glands are evolutionarily newer than vertebral column, so mammary glands are an autapomorphy if vertebral column is an apomorphy, but if mammary glands are the apomorphy being considered then vertebral column is a plesiomorphy.
Relations to other terms
[edit]These phylogenetic terms are used to describe different patterns of ancestral and derived character or trait states as stated in the above diagram in association with apomorphies and synapomorphies.[13][14]
- Symplesiomorphy – an ancestral trait shared by two or more taxa.
- Plesiomorphy – a symplesiomorphy discussed in reference to a more derived state.
- Pseudoplesiomorphy – is a trait that cannot be identified as neither a plesiomorphy nor an apomorphy that is a reversal.[15]
- Reversal – is a loss of derived trait present in ancestor and the reestablishment of a plesiomorphic trait.
- Convergence – independent evolution of a similar trait in two or more taxa.
- Apomorphy – a derived trait. Apomorphy shared by two or more taxa and inherited from a common ancestor is synapomorphy. Apomorphy unique to a given taxon is autapomorphy.[16][17][18][19]
- Synapomorphy/homology – a derived trait that is found in some or all terminal groups of a clade, and inherited from a common ancestor, for which it was an autapomorphy (i.e., not present in its immediate ancestor).
- Underlying synapomorphy – a synapomorphy that has been lost again in many members of the clade. If lost in all but one, it can be hard to distinguish from an autapomorphy.
- Autapomorphy – a distinctive derived trait that is unique to a given taxon or group.[20]
- Homoplasy in biological systematics is when a trait has been gained or lost independently in separate lineages during evolution. This convergent evolution leads to species independently sharing a trait that is different from the trait inferred to have been present in their common ancestor.[21][22][23]
- Parallel homoplasy – derived trait present in two groups or species without a common ancestor due to convergent evolution.[24]
- Reverse homoplasy – trait present in an ancestor but not in direct descendants that reappears in later descendants.[25]
- Hemiplasy is the case where a character that appears homoplastic given the species tree actually has a single origin on the associated gene tree.[26][27] Hemiplasy reflects gene tree-species tree discordance due to the multispecies coalescent.
References
[edit]- ^ a b Roderick D.M. Page; Edward C. Holmes (14 July 2009). Molecular Evolution: A Phylogenetic Approach. John Wiley & Sons. ISBN 978-1-4443-1336-9.
- ^ Futuyma, Douglas J.; Kirkpatrick, Mark (2017). "Tree of life". Evolution (4th ed.). Sunderland, Mass.: Sinauer Associates. pp. 27–53.
- ^ a b Futuyma, Douglas J.; Kirkpatrick, Mark (2017). "Phylogeny: The unity and diversity of life". Evolution (4th ed.). Sunderland, Mass.: Sinauer Associates. pp. 401–429.
- ^ "Reconstructing trees: Cladistics". Understanding Evolution. University of California Museum of Paleontology. 5 May 2021. Retrieved 16 October 2021.
- ^ a b Kitching, Ian J.; Forey, Peter L.; Williams, David M. (2001). "Cladistics". In Levin, Simon A. (ed.). Encyclopedia of Biodiversity (2nd ed.). Elsevier. pp. 33–45. doi:10.1016/B978-0-12-384719-5.00022-8. ISBN 9780123847201. Retrieved 29 August 2021.)
- ^ Hillis, David M.; Sadava, David; Hill, Richard W.; Price, Mary V. (2014). "Reconstructing and using phylogenies". Principles of Life (2nd ed.). Sunderland, Mass.: Sinauer Associates. pp. 325–342. ISBN 978-1464175121.
- ^ Currie PJ, Padia K (1997). Encyclopedia of Dinosaurs. Elsevier. p. 543. ISBN 978-0-08-049474-6.
- ^ Concise Encyclopedia Biology. Tubingen, DEU: Walter de Gruyter. 1996. p. 366. ISBN 9783110106619.
- ^ Barton N, Briggs D, Eisen J, Goldstein D, Patel N (2007). "Phylogenetic Reconstruction". Evolution. Cold Spring Harbor Laboratory Press.
- ^ a b Baum, David (2008). "Trait Evolution on a Phylogenetic Tree: Relatedness, Similarity, and the Myth of Evolutionary Advancement". Nature Education. 1 (1): 191.
- ^ andrewgillis (2016-04-19). "Gills, fins and the evolution of vertebrate paired appendages". the Node. Retrieved 2024-06-09.
- ^ Novick LR, Catley KM. Understanding phylogenies in biology: the influence of a Gestalt perceptual principle. J Exp Psychol Appl. 2007;13:197–223.
- ^ Roderick D.M. Page; Edward C. Holmes (14 July 2009). Molecular Evolution: A Phylogenetic Approach. John Wiley & Sons. ISBN 978-1-4443-1336-9.
- ^ Calow PP (2009). Encyclopedia of Ecology and Environmental Management. John Wiley & Sons. ISBN 978-1-4443-1324-6. OCLC 1039167559.
- ^ Williams D, Schmitt M, Wheeler Q (July 2016). The Future of Phylogenetic Systematics: The Legacy of Willi Hennig. Cambridge University Press. ISBN 978-1-107-11764-8.
- ^ Simpson MG (9 August 2011). Plant Systematics. Amsterdam. ISBN 9780080514048.
{{cite book}}:|work=ignored (help)CS1 maint: location missing publisher (link) - ^ Russell PJ, Hertz PE, McMillan B (2013). Biology: The Dynamic Science. Cengage Learning. ISBN 978-1-285-41534-5.
- ^ Lipscomb D (1998). "Basics of Cladistic Analysis" (PDF). Washington D.C.: George Washington University.
- ^ Choudhuri S (2014-05-09). Bioinformatics for Beginners: Genes, Genomes, Molecular Evolution, Databases and Analytical Tools (1st ed.). Academic Press. p. 51. ISBN 978-0-12-410471-6. OCLC 950546876.
- ^ Appel, Ron D.; Feytmans, Ernest. Bioinformatics: a Swiss Perspective."Chapter 3: Introduction of Phylogenetics and its Molecular Aspects." World Scientific Publishing Company, 1st edition. 2009.
- ^ Gauger A (April 17, 2012). "Similarity Happens! The Problem of Homoplasy". Evolution Today & Science News.
- ^ Sanderson MJ, Hufford L (21 October 1996). Homoplasy: The Recurrence of Similarity in Evolution. Elsevier. ISBN 978-0-08-053411-4. OCLC 173520205.
- ^ Brandley MC, Warren DL, Leaché AD, McGuire JA (April 2009). "Homoplasy and clade support". Systematic Biology. 58 (2): 184–98. doi:10.1093/sysbio/syp019. PMID 20525577.
- ^ Archie JW (September 1989). "Homoplasy Excess Ratios: New Indices for Measuring Levels of Homoplasy in Phylogenetic Systematics and a Critique of the Consistency Index". Systematic Biology. 38 (1): 253–269. doi:10.2307/2992286. JSTOR 2992286.
- ^ Wake DB, Wake MH, Specht CD (February 2011). "Homoplasy: from detecting pattern to determining process and mechanism of evolution". Science. 331 (6020): 1032–5. Bibcode:2011Sci...331.1032W. doi:10.1126/science.1188545. PMID 21350170. S2CID 26845473.
- "Homoplasy: A good thread to pull to understand the evolutionary ball of yarn". ScienceDaily (Press release). February 25, 2011.
- ^ Avise JC, Robinson TJ (June 2008). "Hemiplasy: a new term in the lexicon of phylogenetics". Systematic Biology. 57 (3): 503–7. doi:10.1080/10635150802164587. PMID 18570042.
- ^ Copetti D, Búrquez A, Bustamante E, Charboneau JL, Childs KL, Eguiarte LE, Lee S, Liu TL, McMahon MM, Whiteman NK, Wing RA, Wojciechowski MF, Sanderson MJ (November 2017). "Extensive gene tree discordance and hemiplasy shaped the genomes of North American columnar cacti". Proceedings of the National Academy of Sciences of the United States of America. 114 (45): 12003–12008. Bibcode:2017PNAS..11412003C. doi:10.1073/pnas.1706367114. PMC 5692538. PMID 29078296.
External links
[edit]- Cladistics, Berkeley
