DNA condensation

DNA condensation refers to the process of compacting DNA molecules in vitro or in vivo.[1] Mechanistic details of DNA packing are essential for its functioning in the process of gene regulation in living systems. Condensed DNA often has surprising properties, which one would not predict from classical concepts of dilute solutions. Therefore, DNA condensation in vitro serves as a model system for many processes of physics, biochemistry and biology.[2] In addition, DNA condensation has many potential applications in medicine and biotechnology.[1]
DNA diameter is about 2 nm, while the length of a stretched single molecule may be up to several dozens of centimetres depending on the organism. Many features of the DNA double helix contribute to its large stiffness, including the mechanical properties of the sugar-phosphate backbone, electrostatic repulsion between phosphates (DNA bears on average one elementary negative charge per each 0.17 nm of the double helix), stacking interactions between the bases of each individual strand, and strand-strand interactions. DNA is one of the stiffest natural polymers, yet it is also one of the longest molecules. The persistence length of double-stranded DNA (dsDNA) is a measure of its stiffness or flexibility, which depends on the DNA sequence and the surrounding environment, including factors like salt concentration, pH, and temperature. Under physiological conditions (e.g., near-neutral pH and physiological salt concentrations), the persistence length of dsDNA is generally around 50 nm, which corresponds to approximately 150 base pairs.[1] This means that at large distances DNA can be considered as a flexible rope, and on a short scale as a stiff rod. Like a garden hose, unpacked DNA would randomly occupy a much larger volume than when it is orderly packed. Mathematically, for a non-interacting flexible chain randomly diffusing in 3D, the end-to-end distance would scale as a square root of the polymer length. For real polymers such as DNA, this gives only a very rough estimate; what is important, is that the space available for the DNA in vivo is much smaller than the space that it would occupy in the case of a free diffusion in the solution. To cope with volume constraints, DNA can pack itself in the appropriate solution conditions with the help of ions and other molecules. Usually, DNA condensation is defined as "the collapse of extended DNA chains into compact, orderly particles containing only one or a few molecules".[3] This definition applies to many situations in vitro and is also close to the definition of DNA condensation in bacteria as "adoption of relatively concentrated, compact state occupying a fraction of the volume available".[4] In eukaryotes, the DNA size and the number of other participating players are much larger, and a DNA molecule forms millions of ordered nucleoprotein particles, the nucleosomes, which is just the first of many levels of DNA packing.[1]
In life
[edit]In viruses
[edit]In viruses and bacteriophages, the DNA or RNA is surrounded by a protein capsid, sometimes further enveloped by a lipid membrane. Double-stranded DNA is stored inside the capsid in the form of a spool, which can have different types of coiling leading to different types of liquid-crystalline packing. This packing can change from hexagonal to cholesteric to isotropic at different stages of the phage functioning. Although the double helices are always locally aligned, the DNA inside viruses does not represent real liquid crystals, because it lacks fluidity. On the other hand, DNA condensed in vitro, e.g., with the help of polyamines also present in viruses, is both locally ordered and fluid.[1]
In bacteria
[edit]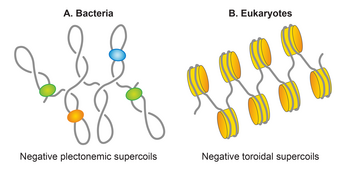
Bacterial DNA is packed with the help of polyamines and proteins called nucleoid-associated proteins. Protein-associated DNA occupies about 1/4 of the intracellular volume forming a concentrated viscous phase with liquid crystalline properties, called the nucleoid. Other research also indicated that the genome of bacteria occupies approximately 10-15% of the bacteria's volume.[5] Similar DNA packaging exists also in chloroplasts and mitochondria. Bacterial DNA is sometimes referred to as the bacterial chromosome. Bacterial nucleoid evolutionary represents an intermediate engineering solution between the protein-free DNA packing in viruses and protein-determined packing in eukaryotes.[1]
Sister chromosomes in the bacterium Escherichia coli are induced by stressful conditions to condense and undergo pairing.[6] Stress-induced condensation occurs by a non-random, zipper-like convergence of sister chromosomes. This convergence appears to depend on the ability of identical double-stranded DNA molecules to specifically identify each other, a process that culminates in the proximity of homologous sites along the paired chromosomes. Diverse stress conditions appear to prime bacteria to effectively cope with severe DNA damages such as double-strand breaks. The apposition of homologous sites associated with stress-induced chromosome condensation helps explain how repair of double-strand breaks and other damages occurs.[6]
In eukaryotes
[edit]
Eukaryotic DNA with a typical length of dozens of centimeters should be orderly packed to be readily accessible inside the micrometer-size nucleus. In most eukaryotes, DNA is arranged in the cell nucleus with the help of histones. In this case, the basic level of DNA compaction is the nucleosome, where the double helix is wrapped around the histone octamer containing two copies of each histone H2A, H2B, H3 and H4. Linker histone H1 binds the DNA between nucleosomes and facilitates packaging of the 10 nm "beads on the string" nucleosomal chain into a more condensed 30 nm fiber. Most of the time, between cell divisions, chromatin is optimized to allow easy access of transcription factors to active genes, which are characterized by a less compact structure called euchromatin, and to alleviate protein access in more tightly packed regions called heterochromatin. During the cell division, chromatin compaction increases even more to form chromosomes, which can cope with large mechanical forces dragging them into each of the two daughter cells.[1] Many aspects of transcription are controlled by chemical modification on the histone proteins, known as the histone code.
Chromosome scaffold has important role to hold the chromatin into compact chromosome. Chromosome scaffold is made of proteins including condensin, topoisomerase IIα and kinesin family member 4 (KIF4)[7]
Dinoflagellates are very divergent eukaryotes in terms of how they package their DNA. Their chromosomes are packed in a liquid-crystalline state.[8] They have lost many of the conserved histone genes, using mostly dinoflagellate viral nucleoproteins (DVNPs) or bacteria-derived dinoflagellate histone-like proteins (HLPs) for packaging instead. It is unknown how they control access to genes; those do retain histone have a special histone code.[9][10]
In archaea
[edit]Depending on the organism, an archaeon may use a bacteria-like HU system or a eukaryote-like nucleosome system for packaging.[11]
In vitro
[edit]DNA condensation can be induced in vitro either by applying external force to bring the double helices together, or by inducing attractive interactions between the DNA segments. The former can be achieved e.g. with the help of the osmotic pressure exerted by crowding neutral polymers in the presence of monovalent salts. In this case, the forces pushing the double helices together are coming from entropic random collisions with the crowding polymers surrounding DNA condensates, and salt is required to neutralize DNA charges and decrease DNA-DNA repulsion. The second possibility can be realized by inducing attractive interactions between the DNA segments by multivalent cationic charged ligands (multivalent metal ions, inorganic cations, polyamines, protamines, peptides, lipids, liposomes and proteins).[1]
Physics
[edit]Condensation of long double-helical DNAs is a sharp phase transition, which takes place within a narrow interval of condensing agent concentrations.[ref] Since the double helices come very closely to each other in the condensed phase, this leads to the restructuring of water molecules, which gives rise to the so-called hydration forces.[ref] To understand attraction between negatively charged DNA molecules, one also must account for correlations between counterions in the solution.[ref] DNA condensation by proteins can exhibit hysteresis, which can be explained using a modified Ising model.[12]
Role in gene regulation
[edit]Nowadays descriptions of gene regulation are based on the approximations of equilibrium binding in dilute solutions, although it is clear that these assumptions are in fact violated in chromatin. The dilute-solution approximation is violated for two reasons. First, the chromatin content is far from being dilute, and second, the numbers of the participating molecules are sometimes so small, that it does not make sense to talk about the bulk concentrations. Further differences from dilute solutions arise due to the different binding affinities of proteins to condensed and uncondensed DNA. Thus in condensed DNA both the reaction rates can be changed and their dependence on the concentrations of reactants may become nonlinear.[1]
See also
[edit]References
[edit]- ^ a b c d e f g h i Teif, VB; Bohinc, K (2011). "Condensed DNA: condensing the concepts". Progress in Biophysics and Molecular Biology. 105 (3): 208–22. doi:10.1016/j.pbiomolbio.2010.07.002. PMID 20638406.
- ^ Bloomfield, VA (1996). "DNA condensation". Current Opinion in Structural Biology. 6 (3): 334–41. doi:10.1016/S0959-440X(96)80052-2. PMID 8804837.
- ^ Bloomfield, VA (1997). "DNA condensation by multivalent cations". Biopolymers. 44 (3): 269–82. CiteSeerX 10.1.1.475.3765. doi:10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. PMID 9591479.
- ^ Zimmerman, SB; Murphy, LD (1996). "Macromolecular crowding and the mandatory condensation of DNA in bacteria". FEBS Letters. 390 (3): 245–8. doi:10.1016/0014-5793(96)00725-9. PMID 8706869.
- ^ Dame, Remus T.; Tark-Dame, Mariliis (June 2016). "Bacterial chromatin: converging views at different scales". Current Opinion in Cell Biology. 40: 60–65. doi:10.1016/j.ceb.2016.02.015. ISSN 1879-0410. PMID 26942688.
- ^ a b Shechter N, Zaltzman L, Weiner A, Brumfeld V, Shimoni E, Fridmann-Sirkis Y, Minsky A (2013). "Stress-induced condensation of bacterial genomes results in re-pairing of sister chromosomes: implications for double strand DNA break repair". J. Biol. Chem. 288 (35): 25659–67. doi:10.1074/jbc.M113.473025. PMC 3757227. PMID 23884460.
- ^ Chromosome Scaffold is a Double-Stranded Assembly of Scaffold Proteins, by Poonperm et al, Nature scientific reports
- ^ Chow, MH; Yan, KTH; Bennett, MJ; Wong, JTY (2010). "Birefringence and DNA condensation of liquid crystalline chromosomes". Eukaryotic Cell. 9 (10): 1577–87. doi:10.1128/EC.00026-10. PMC 2950428. PMID 20400466.
- ^ Marinov GK, Lynch M (2016). "Diversity and Divergence of Dinoflagellate Histone Proteins". G3 (Bethesda). 6 (2): 397–422. doi:10.1534/g3.115.023275. PMC 4751559. PMID 26646152.
- ^ Riaz, S; Sui, Z; Niaz, Z; Khan, S; Liu, Y; Liu, H (14 December 2018). "Distinctive Nuclear Features of Dinoflagellates with A Particular Focus on Histone and Histone-Replacement Proteins". Microorganisms. 6 (4): 128. doi:10.3390/microorganisms6040128. PMC 6313786. PMID 30558155.
- ^ Luijsterburg, Martijn S.; White, Malcolm F.; van Driel, Roel; Dame, Remus Th. (8 January 2009). "The Major Architects of Chromatin: Architectural Proteins in Bacteria, Archaea and Eukaryotes". Critical Reviews in Biochemistry and Molecular Biology. 43 (6): 393–418. doi:10.1080/10409230802528488. PMID 19037758. S2CID 85874882.
- ^ Vtyurina, Natalia N.; Dulin, David; Docter, Margreet W.; Meyer, Anne S.; Dekker, Nynke H.; Abbondanzieri, Elio A. (2016-04-18). "Hysteresis in DNA compaction by Dps is described by an Ising model". Proceedings of the National Academy of Sciences. 113 (18): 4982–4987. Bibcode:2016PNAS..113.4982V. doi:10.1073/pnas.1521241113. ISSN 0027-8424. PMC 4983820. PMID 27091987.
Further reading
[edit]- Gelbart W. M.; Bruinsma R.; Pincus P. A.; Parsegian V. A. (2000). "DNA-Inspired Electrostatics". Physics Today. 53 (9): 38. Bibcode:2000PhT....53i..38G. doi:10.1063/1.1325230.
- Strey H. H.; Podgornik R.; Rau D. C.; Parsegian V. A. (1998). "DNA-DNA interactions". Current Opinion in Structural Biology. 8 (3): 309–313. doi:10.1016/s0959-440x(98)80063-8. PMID 9666326.
- Schiessel H (2003). "The physics of chromatin". J. Phys.: Condens. Matter. 15 (19): R699–R774. arXiv:cond-mat/0303455. Bibcode:2003JPCM...15R.699S. doi:10.1088/0953-8984/15/19/203. S2CID 250893202.
- Vijayanathan V.; Thomas T.; Thomas T. J. (2002). "DNA nanoparticles and development of DNA delivery vehicles for gene therapy". Biochemistry. 41 (48): 14085–14094. doi:10.1021/bi0203987. PMID 12450371.
- Yoshikawa K (2001). "Controlling the higher-order structure of giant DNA molecules". Advanced Drug Delivery Reviews. 52 (3): 235–244. doi:10.1016/s0169-409x(01)00210-1. PMID 11718948.
- Hud N. V.; Vilfan I. D. (2005). "Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size". Annu Rev Biophys Biomol Struct. 34: 295–318. doi:10.1146/annurev.biophys.34.040204.144500. PMID 15869392.
- Yoshikawa, K., and Y. Yoshikawa. 2002. Compaction and condensation of DNA. In Pharmaceutical perspectives of nucleic acid-based therapeutics. R. I. Mahato, and S. W. Kim, editors. Taylor & Francis. 137–163.
